Please be patient as the page loads
|
CT194_HUMAN
|
||||||
| SwissProt Accessions | Q5TEA3, Q66K86, Q6P2R9, Q9UFX9 | Gene names | C20orf194 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf194. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CT194_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TEA3, Q66K86, Q6P2R9, Q9UFX9 | Gene names | C20orf194 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf194. | |||||
|
CT194_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995936 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT23, Q3UPD4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf194 homolog. | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 3) | NC score | 0.024395 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
CHL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.019556 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00533, Q2M3G2, Q59FY0 | Gene names | CHL1, CALL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Close homolog of L1). | |||||
|
NUDT9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 5) | NC score | 0.046815 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
RAE1D_MOUSE
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.035357 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI58 | Gene names | Raet1d | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid early inducible protein 1 delta precursor (RAE-1delta). | |||||
|
CHL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.014321 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.041744 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
OSCP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.025872 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVF1, Q4AEJ0, Q8N7G2, Q8TDF1 | Gene names | OSCP1, C1orf102, NOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OSCP1 (Organic solute transport protein 1) (hOSCP1) (Oxidored- nitro domain-containing protein 1). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.017556 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
KPCZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.001502 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05513, Q15207, Q5SYT5, Q969S4 | Gene names | PRKCZ, PKC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C zeta type (EC 2.7.11.13) (nPKC-zeta). | |||||
|
RAE1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.026045 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08603 | Gene names | Raet1b | |||
|
Domain Architecture |
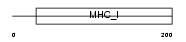
|
|||||
| Description | Retinoic acid early inducible protein 1 beta precursor (RAE-1beta). | |||||
|
CXB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.006678 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08034 | Gene names | GJB1, CX32 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction beta-1 protein (Connexin-32) (Cx32) (GAP junction 28 kDa liver protein). | |||||
|
SRP72_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.022565 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.006573 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.006343 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.008014 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
CT194_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TEA3, Q66K86, Q6P2R9, Q9UFX9 | Gene names | C20orf194 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf194. | |||||
|
CT194_MOUSE
|
||||||
| NC score | 0.995936 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT23, Q3UPD4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf194 homolog. | |||||
|
NUDT9_HUMAN
|
||||||
| NC score | 0.046815 (rank : 3) | θ value | 1.06291 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.041744 (rank : 4) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
RAE1D_MOUSE
|
||||||
| NC score | 0.035357 (rank : 5) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI58 | Gene names | Raet1d | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid early inducible protein 1 delta precursor (RAE-1delta). | |||||
|
RAE1B_MOUSE
|
||||||
| NC score | 0.026045 (rank : 6) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08603 | Gene names | Raet1b | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid early inducible protein 1 beta precursor (RAE-1beta). | |||||
|
OSCP1_HUMAN
|
||||||
| NC score | 0.025872 (rank : 7) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVF1, Q4AEJ0, Q8N7G2, Q8TDF1 | Gene names | OSCP1, C1orf102, NOR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OSCP1 (Organic solute transport protein 1) (hOSCP1) (Oxidored- nitro domain-containing protein 1). | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.024395 (rank : 8) | θ value | 0.365318 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
SRP72_HUMAN
|
||||||
| NC score | 0.022565 (rank : 9) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
CHL1_HUMAN
|
||||||
| NC score | 0.019556 (rank : 10) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00533, Q2M3G2, Q59FY0 | Gene names | CHL1, CALL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Close homolog of L1). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.017556 (rank : 11) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
CHL1_MOUSE
|
||||||
| NC score | 0.014321 (rank : 12) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70232, Q8BS24, Q8C6W0, Q8C823, Q8VBY7 | Gene names | Chl1, Call | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural cell adhesion molecule L1-like protein precursor (Cell adhesion molecule with homology to L1CAM) (Close homolog of L1) (Chl1-like protein). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.008014 (rank : 13) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
CXB1_HUMAN
|
||||||
| NC score | 0.006678 (rank : 14) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08034 | Gene names | GJB1, CX32 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction beta-1 protein (Connexin-32) (Cx32) (GAP junction 28 kDa liver protein). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.006573 (rank : 15) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.006343 (rank : 16) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
KPCZ_HUMAN
|
||||||
| NC score | 0.001502 (rank : 17) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05513, Q15207, Q5SYT5, Q969S4 | Gene names | PRKCZ, PKC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C zeta type (EC 2.7.11.13) (nPKC-zeta). | |||||