Please be patient as the page loads
|
CM35A_HUMAN
|
||||||
| SwissProt Accessions | Q08708 | Gene names | CD300C, CMRF35, CMRF35A | |||
|
Domain Architecture |
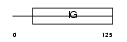
|
|||||
| Description | CMRF35-A antigen precursor (CMRF-35) (CD300c antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CM35A_HUMAN
|
||||||
| θ value | 4.47364e-108 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q08708 | Gene names | CD300C, CMRF35, CMRF35A | |||
|
Domain Architecture |

|
|||||
| Description | CMRF35-A antigen precursor (CMRF-35) (CD300c antigen). | |||||
|
CM35H_HUMAN
|
||||||
| θ value | 1.01059e-59 (rank : 2) | NC score | 0.936945 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UGN4, O95100, Q9HD97, Q9UBK4, Q9UMS9, Q9UMT0 | Gene names | CD300A, CMRF35H | |||
|
Domain Architecture |
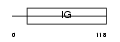
|
|||||
| Description | CMRF35-H antigen precursor (CMRF35-H9) (CMRF-35-H9) (CD300a antigen) (Inhibitory receptor protein 60) (IRp60) (IRC1/IRC2) (NK inhibitory receptor). | |||||
|
CLM1_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 3) | NC score | 0.736974 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6SJQ7, Q6PEU7, Q8K4V9 | Gene names | Cd300lf, Clm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (CD300 antigen like family member F). | |||||
|
CLM1_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 4) | NC score | 0.724593 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDQ1, Q3Y6P0, Q6UX24, Q7Z6A6, Q7Z7I4, Q7Z7I5, Q8N6D0, Q8NAF5 | Gene names | CD300LF, CLM1, IGSF13, IREM1, NKIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (Immune receptor expressed on myeloid cells protein 1) (IREM-1) (Immunoglobulin superfamily member 13) (NK inhibitory receptor) (CD300 antigen like family member F) (IgSF13). | |||||
|
PIGR_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 5) | NC score | 0.528738 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01833, Q8IZY7 | Gene names | PIGR | |||
|
Domain Architecture |

|
|||||
| Description | Polymeric-immunoglobulin receptor precursor (Poly-Ig receptor) (PIGR) (Hepatocellular carcinoma-associated protein TB6) [Contains: Secretory component]. | |||||
|
PIGR_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 6) | NC score | 0.498384 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70570 | Gene names | Pigr | |||
|
Domain Architecture |

|
|||||
| Description | Polymeric-immunoglobulin receptor precursor (Poly-Ig receptor) (PIGR) [Contains: Secretory component]. | |||||
|
NCTR2_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 7) | NC score | 0.573515 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |
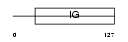
|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
TRML2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 8) | NC score | 0.400991 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2LA85 | Gene names | Treml2, Tlt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
TRML1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 9) | NC score | 0.358093 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K558, Q9D3N6 | Gene names | Treml1, Tlt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
TREM2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 10) | NC score | 0.445415 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NH8, Q8CGK4, Q8CIA6, Q99NH9, Q9JL34 | Gene names | Trem2, Trem2a, Trem2b, Trem2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 2 precursor (Triggering receptor expressed on monocytes 2) (TREM-2). | |||||
|
TRML1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.391471 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86YW5, Q496B3, Q8IWY1, Q8IWY2 | Gene names | TREML1, TLT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
TREM2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 12) | NC score | 0.407029 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZC2, Q8N5H8, Q8WYN6 | Gene names | TREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 2 precursor (Triggering receptor expressed on monocytes 2) (TREM-2). | |||||
|
VSIG2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.122625 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z109, Q9CVA4, Q9D0T4 | Gene names | Vsig2, Ctm, Ctxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | V-set and immunoglobulin domain-containing protein 2 precursor (CT- like protein) (Cortical thymocyte-like protein). | |||||
|
TIMD1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.272890 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5QNS5, Q7TPU2, Q8VIM1, Q8VIM2 | Gene names | Havcr1, Tim1, Timd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 homolog precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1). | |||||
|
VSIG2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 15) | NC score | 0.114605 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96IQ7, O95791, Q9NX42 | Gene names | VSIG2, CTH, CTXL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | V-set and immunoglobulin domain-containing protein 2 precursor (CT- like protein) (Cortical thymocyte-like protein). | |||||
|
TRML2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 16) | NC score | 0.372246 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T2D2, Q8IWY0, Q9H8E9 | Gene names | TREML2, C6orf76, TLT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
TYRO3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.030218 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55144, O09070, O09080, P70285, Q60752, Q62482, Q62483, Q62484, Q78E85, Q78E87 | Gene names | Tyro3, Dtk, Rse | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase DTK) (TK19-2) (Etk2/tyro3). | |||||
|
TIMD2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.228469 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R183, Q8VBW0 | Gene names | Timd2, Tim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 2 precursor (TIMD-2) (T cell membrane protein 2) (TIM-2). | |||||
|
TIMD4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.222426 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96H15 | Gene names | TIMD4, TIM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 4 precursor (TIMD-4) (T-cell membrane protein 4) (TIM-4). | |||||
|
TYRO3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.024260 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06418, O14953, Q86VR3 | Gene names | TYRO3, BYK, DTK, RSE, SKY | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase SKY) (Tyrosine- protein kinase DTK) (Protein-tyrosine kinase byk). | |||||
|
KV2F_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.053349 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06310 | Gene names | ||||
|
Domain Architecture |
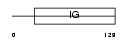
|
|||||
| Description | Ig kappa chain V-II region RPMI 6410 precursor. | |||||
|
LY9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.090286 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01965, Q9ES29, Q9ES35, Q9ES36 | Gene names | Ly9, Ly-9 | |||
|
Domain Architecture |
|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9). | |||||
|
CD7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.113798 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09564 | Gene names | CD7 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell antigen CD7 precursor (GP40) (T-cell leukemia antigen) (TP41) (Leu-9). | |||||
|
CEAM1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.072419 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13688, O60430, Q13858, Q15600, Q9UQV9 | Gene names | CEACAM1, BGP, BGP1 | |||
|
Domain Architecture |
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (CD66 antigen) (CD66a antigen). | |||||
|
IZUM1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.080206 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
KV1F_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.048526 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P01598 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region EU. | |||||
|
TIMD1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.216409 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
TIMD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.178439 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6U7R4, Q6U7R3, Q8CIC7 | Gene names | Timd4, Tim4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 4 precursor (TIMD-4) (T-cell membrane protein 4) (TIM-4) (Spleen, mucin- containing, knockout of lymphotoxin protein) (SMUCKLER). | |||||
|
CEAM5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.062737 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
TIMD3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.169900 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VIM0 | Gene names | Havcr2, Tim3, Timd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 2 homolog precursor (HAVcr-2) (T cell immunoglobulin and mucin domain-containing protein 3) (TIMD-3) (T cell membrane protein 3) (TIM-3). | |||||
|
KV2A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.051606 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
KV5P_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.045420 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01649 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-V regions (Anti-arsonate antibodies). | |||||
|
SHPS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.085616 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97797, O08907, O35924, O88555, O88556, P97796, Q8R559, Q9QX57, Q9WTN4 | Gene names | Sirpa, Bit, Myd1, Ptpns1, Shps1, Sirp | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (mSIRP-alpha1) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (p84) (CD172a antigen). | |||||
|
KV1E_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.046896 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01597 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region DEE. | |||||
|
KV1Y_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.046968 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80362 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region WAT. | |||||
|
KV2B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.058324 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |
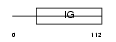
|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
SIRB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.069793 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
Domain Architecture |
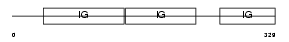
|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
CEAM8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.065187 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |
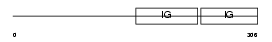
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||
|
HPLN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.031832 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |
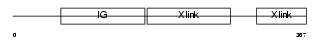
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
KV1D_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.046801 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01596 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region CAR. | |||||
|
KV1W_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.056737 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04431 | Gene names | ||||
|
Domain Architecture |
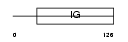
|
|||||
| Description | Ig kappa chain V-I region Walker precursor. | |||||
|
SHPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.069348 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78324, O00683, O43799, Q8N517, Q8TAL8, Q9H0Z2, Q9UDX2, Q9UIJ6, Q9Y4U9 | Gene names | SIRPA, BIT, MFR, MYD1, PTPNS1, SHPS1, SIRP | |||
|
Domain Architecture |
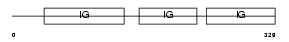
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (Sirp-alpha-2) (Sirp-alpha- 3) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (Macrophage fusion receptor) (p84) (CD172a antigen). | |||||
|
FCRLA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.025759 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.031614 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
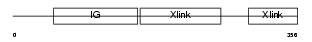
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
JAM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.090696 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57087, Q6UXG6, Q6YNC1 | Gene names | JAM2, C21orf43, VEJAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
JAM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.089355 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI59, Q8C5K9, Q8CE95 | Gene names | Jam2, Vejam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
KV1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.045844 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01604 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Kue. | |||||
|
LY9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.059491 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HBG7, Q14775, Q5VYI3, Q9H4N5, Q9NQ24 | Gene names | LY9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9) (CD229 antigen). | |||||
|
HV46_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.016887 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P01822 | Gene names | ||||
|
Domain Architecture |
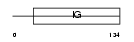
|
|||||
| Description | Ig heavy chain V region MOPC 315 precursor. | |||||
|
IGSF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.043634 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q93033, Q15856 | Gene names | IGSF2, CD101, EWI101, V7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 2 precursor (Glu-Trp-Ile EWI motif- containing protein 101) (EWI-101) (Cell surface glycoprotein V7) (CD101 antigen). | |||||
|
CD226_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.052382 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15762 | Gene names | CD226, DNAM1 | |||
|
Domain Architecture |
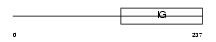
|
|||||
| Description | CD226 antigen precursor (DNAX accessory molecule 1) (DNAM-1). | |||||
|
FGRL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.036474 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
KV1G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.044596 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P01599 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Gal. | |||||
|
KV1Q_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.046825 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01609 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Scw. | |||||
|
TREM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.131118 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NP99, Q53FL8, Q5T2C9, Q86YU1 | Gene names | TREM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 1 precursor (TREM-1) (Triggering receptor expressed on monocytes 1). | |||||
|
GI24_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.041112 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D659, Q6KAT9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet receptor Gi24 precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.006391 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TIMD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.158461 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDQ0, Q8WW60, Q96K94 | Gene names | HAVCR2, TIM3, TIMD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 2 precursor (HAVcr-2) (T cell immunoglobulin and mucin domain-containing protein 3) (TIMD-3) (T cell membrane protein 3) (TIM-3). | |||||
|
CD22_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.051663 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
CEAM6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.064672 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40199, Q14920 | Gene names | CEACAM6, NCA | |||
|
Domain Architecture |
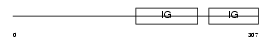
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 6 precursor (Normal cross-reacting antigen) (Nonspecific crossreacting antigen) (CD66c antigen). | |||||
|
HV12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.009445 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01756 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig heavy chain V region MOPC 104E. | |||||
|
KV1P_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.044426 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01608 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Roy. | |||||
|
KV5R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.041564 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01651 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-V region EPC 109. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.005185 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SCN2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.066302 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q56A07 | Gene names | Scn2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel beta-2 subunit precursor. | |||||
|
CD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.056601 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50283 | Gene names | Cd7 | |||
|
Domain Architecture |
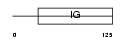
|
|||||
| Description | T-cell antigen CD7 precursor. | |||||
|
CXAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.053263 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |
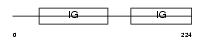
|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
CXAR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.061064 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97792, O09052, Q3ULD0, Q91W66, Q99KG0, Q9DBJ8 | Gene names | Cxadr, Car | |||
|
Domain Architecture |
|
|||||
| Description | Coxsackievirus and adenovirus receptor homolog precursor (CAR) (mCAR). | |||||
|
ESAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.057554 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
ESAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.062377 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
GPA33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.089555 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99795 | Gene names | GPA33 | |||
|
Domain Architecture |
|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33). | |||||
|
GPA33_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.087118 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JKA5, Q922D5 | Gene names | Gpa33 | |||
|
Domain Architecture |
|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33) (mA33). | |||||
|
JAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.076393 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y624 | Gene names | F11R, JAM1, JCAM | |||
|
Domain Architecture |
|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (Platelet adhesion molecule 1) (PAM-1) (Platelet F11 receptor) (CD321 antigen). | |||||
|
JAM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.067869 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88792 | Gene names | F11r, Jam1, Jcam, Jcam1 | |||
|
Domain Architecture |
|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (CD321 antigen). | |||||
|
JAM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050944 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8B7, Q8BT59, Q9D1M9, Q9EPK4 | Gene names | Jam3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctional adhesion molecule C precursor (JAM-C) (Junctional adhesion molecule 3) (JAM-3) (JAM-2). | |||||
|
SIRPD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.055992 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H106, Q5TFQ6 | Gene names | SIRPD, PTPNS1L2 | |||
|
Domain Architecture |
|
|||||
| Description | Signal-regulatory protein delta precursor (Protein tyrosine phosphatase non-receptor type substrate 1-like 2). | |||||
|
TREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.132236 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKE2, Q3TAL7 | Gene names | Trem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 1 precursor (TREM-1). | |||||
|
CM35A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.47364e-108 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q08708 | Gene names | CD300C, CMRF35, CMRF35A | |||
|
Domain Architecture |

|
|||||
| Description | CMRF35-A antigen precursor (CMRF-35) (CD300c antigen). | |||||
|
CM35H_HUMAN
|
||||||
| NC score | 0.936945 (rank : 2) | θ value | 1.01059e-59 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UGN4, O95100, Q9HD97, Q9UBK4, Q9UMS9, Q9UMT0 | Gene names | CD300A, CMRF35H | |||
|
Domain Architecture |

|
|||||
| Description | CMRF35-H antigen precursor (CMRF35-H9) (CMRF-35-H9) (CD300a antigen) (Inhibitory receptor protein 60) (IRp60) (IRC1/IRC2) (NK inhibitory receptor). | |||||
|
CLM1_MOUSE
|
||||||
| NC score | 0.736974 (rank : 3) | θ value | 1.38178e-16 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6SJQ7, Q6PEU7, Q8K4V9 | Gene names | Cd300lf, Clm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (CD300 antigen like family member F). | |||||
|
CLM1_HUMAN
|
||||||
| NC score | 0.724593 (rank : 4) | θ value | 1.80466e-16 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDQ1, Q3Y6P0, Q6UX24, Q7Z6A6, Q7Z7I4, Q7Z7I5, Q8N6D0, Q8NAF5 | Gene names | CD300LF, CLM1, IGSF13, IREM1, NKIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMRF35-like-molecule 1 precursor (CLM-1) (Immune receptor expressed on myeloid cells protein 1) (IREM-1) (Immunoglobulin superfamily member 13) (NK inhibitory receptor) (CD300 antigen like family member F) (IgSF13). | |||||
|
NCTR2_HUMAN
|
||||||
| NC score | 0.573515 (rank : 5) | θ value | 4.45536e-07 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |

|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
PIGR_HUMAN
|
||||||
| NC score | 0.528738 (rank : 6) | θ value | 2.69671e-12 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01833, Q8IZY7 | Gene names | PIGR | |||
|
Domain Architecture |

|
|||||
| Description | Polymeric-immunoglobulin receptor precursor (Poly-Ig receptor) (PIGR) (Hepatocellular carcinoma-associated protein TB6) [Contains: Secretory component]. | |||||
|
PIGR_MOUSE
|
||||||
| NC score | 0.498384 (rank : 7) | θ value | 2.79066e-09 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70570 | Gene names | Pigr | |||
|
Domain Architecture |

|
|||||
| Description | Polymeric-immunoglobulin receptor precursor (Poly-Ig receptor) (PIGR) [Contains: Secretory component]. | |||||
|
TREM2_MOUSE
|
||||||
| NC score | 0.445415 (rank : 8) | θ value | 4.1701e-05 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NH8, Q8CGK4, Q8CIA6, Q99NH9, Q9JL34 | Gene names | Trem2, Trem2a, Trem2b, Trem2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 2 precursor (Triggering receptor expressed on monocytes 2) (TREM-2). | |||||
|
TREM2_HUMAN
|
||||||
| NC score | 0.407029 (rank : 9) | θ value | 0.000270298 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZC2, Q8N5H8, Q8WYN6 | Gene names | TREM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 2 precursor (Triggering receptor expressed on monocytes 2) (TREM-2). | |||||
|
TRML2_MOUSE
|
||||||
| NC score | 0.400991 (rank : 10) | θ value | 2.44474e-05 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2LA85 | Gene names | Treml2, Tlt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
TRML1_HUMAN
|
||||||
| NC score | 0.391471 (rank : 11) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86YW5, Q496B3, Q8IWY1, Q8IWY2 | Gene names | TREML1, TLT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
TRML2_HUMAN
|
||||||
| NC score | 0.372246 (rank : 12) | θ value | 0.00869519 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5T2D2, Q8IWY0, Q9H8E9 | Gene names | TREML2, C6orf76, TLT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 2 protein precursor (TLT-2) (Triggering receptor expressed on myeloid cells-like protein 2). | |||||
|
TRML1_MOUSE
|
||||||
| NC score | 0.358093 (rank : 13) | θ value | 3.19293e-05 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K558, Q9D3N6 | Gene names | Treml1, Tlt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
TIMD1_MOUSE
|
||||||
| NC score | 0.272890 (rank : 14) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5QNS5, Q7TPU2, Q8VIM1, Q8VIM2 | Gene names | Havcr1, Tim1, Timd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 homolog precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1). | |||||
|
TIMD2_MOUSE
|
||||||
| NC score | 0.228469 (rank : 15) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R183, Q8VBW0 | Gene names | Timd2, Tim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 2 precursor (TIMD-2) (T cell membrane protein 2) (TIM-2). | |||||
|
TIMD4_HUMAN
|
||||||
| NC score | 0.222426 (rank : 16) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96H15 | Gene names | TIMD4, TIM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 4 precursor (TIMD-4) (T-cell membrane protein 4) (TIM-4). | |||||
|
TIMD1_HUMAN
|
||||||
| NC score | 0.216409 (rank : 17) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
TIMD4_MOUSE
|
||||||
| NC score | 0.178439 (rank : 18) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6U7R4, Q6U7R3, Q8CIC7 | Gene names | Timd4, Tim4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell immunoglobulin and mucin domain-containing protein 4 precursor (TIMD-4) (T-cell membrane protein 4) (TIM-4) (Spleen, mucin- containing, knockout of lymphotoxin protein) (SMUCKLER). | |||||
|
TIMD3_MOUSE
|
||||||
| NC score | 0.169900 (rank : 19) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VIM0 | Gene names | Havcr2, Tim3, Timd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 2 homolog precursor (HAVcr-2) (T cell immunoglobulin and mucin domain-containing protein 3) (TIMD-3) (T cell membrane protein 3) (TIM-3). | |||||
|
TIMD3_HUMAN
|
||||||
| NC score | 0.158461 (rank : 20) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDQ0, Q8WW60, Q96K94 | Gene names | HAVCR2, TIM3, TIMD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 2 precursor (HAVcr-2) (T cell immunoglobulin and mucin domain-containing protein 3) (TIMD-3) (T cell membrane protein 3) (TIM-3). | |||||
|
TREM1_MOUSE
|
||||||
| NC score | 0.132236 (rank : 21) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKE2, Q3TAL7 | Gene names | Trem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 1 precursor (TREM-1). | |||||
|
TREM1_HUMAN
|
||||||
| NC score | 0.131118 (rank : 22) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NP99, Q53FL8, Q5T2C9, Q86YU1 | Gene names | TREM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Triggering receptor expressed on myeloid cells 1 precursor (TREM-1) (Triggering receptor expressed on monocytes 1). | |||||
|
VSIG2_MOUSE
|
||||||
| NC score | 0.122625 (rank : 23) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z109, Q9CVA4, Q9D0T4 | Gene names | Vsig2, Ctm, Ctxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | V-set and immunoglobulin domain-containing protein 2 precursor (CT- like protein) (Cortical thymocyte-like protein). | |||||
|
VSIG2_HUMAN
|
||||||
| NC score | 0.114605 (rank : 24) | θ value | 0.00390308 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96IQ7, O95791, Q9NX42 | Gene names | VSIG2, CTH, CTXL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | V-set and immunoglobulin domain-containing protein 2 precursor (CT- like protein) (Cortical thymocyte-like protein). | |||||
|
CD7_HUMAN
|
||||||
| NC score | 0.113798 (rank : 25) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09564 | Gene names | CD7 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell antigen CD7 precursor (GP40) (T-cell leukemia antigen) (TP41) (Leu-9). | |||||
|
JAM2_HUMAN
|
||||||
| NC score | 0.090696 (rank : 26) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P57087, Q6UXG6, Q6YNC1 | Gene names | JAM2, C21orf43, VEJAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
LY9_MOUSE
|
||||||
| NC score | 0.090286 (rank : 27) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01965, Q9ES29, Q9ES35, Q9ES36 | Gene names | Ly9, Ly-9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9). | |||||
|
GPA33_HUMAN
|
||||||
| NC score | 0.089555 (rank : 28) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99795 | Gene names | GPA33 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33). | |||||
|
JAM2_MOUSE
|
||||||
| NC score | 0.089355 (rank : 29) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI59, Q8C5K9, Q8CE95 | Gene names | Jam2, Vejam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
GPA33_MOUSE
|
||||||
| NC score | 0.087118 (rank : 30) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JKA5, Q922D5 | Gene names | Gpa33 | |||
|
Domain Architecture |

|
|||||
| Description | Cell surface A33 antigen precursor (Glycoprotein A33) (mA33). | |||||
|
SHPS1_MOUSE
|
||||||
| NC score | 0.085616 (rank : 31) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97797, O08907, O35924, O88555, O88556, P97796, Q8R559, Q9QX57, Q9WTN4 | Gene names | Sirpa, Bit, Myd1, Ptpns1, Shps1, Sirp | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (mSIRP-alpha1) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (p84) (CD172a antigen). | |||||
|
IZUM1_MOUSE
|
||||||
| NC score | 0.080206 (rank : 32) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D9J7 | Gene names | Izumo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Izumo sperm-egg fusion protein 1 precursor (Sperm-specific protein izumo). | |||||
|
JAM1_HUMAN
|
||||||
| NC score | 0.076393 (rank : 33) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y624 | Gene names | F11R, JAM1, JCAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (Platelet adhesion molecule 1) (PAM-1) (Platelet F11 receptor) (CD321 antigen). | |||||
|
CEAM1_HUMAN
|
||||||
| NC score | 0.072419 (rank : 34) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13688, O60430, Q13858, Q15600, Q9UQV9 | Gene names | CEACAM1, BGP, BGP1 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 1 precursor (Biliary glycoprotein 1) (BGP-1) (CD66 antigen) (CD66a antigen). | |||||
|
SIRB1_HUMAN
|
||||||
| NC score | 0.069793 (rank : 35) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
SHPS1_HUMAN
|
||||||
| NC score | 0.069348 (rank : 36) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78324, O00683, O43799, Q8N517, Q8TAL8, Q9H0Z2, Q9UDX2, Q9UIJ6, Q9Y4U9 | Gene names | SIRPA, BIT, MFR, MYD1, PTPNS1, SHPS1, SIRP | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (Sirp-alpha-2) (Sirp-alpha- 3) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (Macrophage fusion receptor) (p84) (CD172a antigen). | |||||
|
JAM1_MOUSE
|
||||||
| NC score | 0.067869 (rank : 37) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88792 | Gene names | F11r, Jam1, Jcam, Jcam1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule A precursor (JAM-A) (Junctional adhesion molecule 1) (JAM-1) (CD321 antigen). | |||||
|
SCN2B_MOUSE
|
||||||
| NC score | 0.066302 (rank : 38) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q56A07 | Gene names | Scn2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel beta-2 subunit precursor. | |||||
|
CEAM8_HUMAN
|
||||||
| NC score | 0.065187 (rank : 39) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||
|
CEAM6_HUMAN
|
||||||
| NC score | 0.064672 (rank : 40) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40199, Q14920 | Gene names | CEACAM6, NCA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 6 precursor (Normal cross-reacting antigen) (Nonspecific crossreacting antigen) (CD66c antigen). | |||||
|
CEAM5_HUMAN
|
||||||
| NC score | 0.062737 (rank : 41) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06731 | Gene names | CEACAM5, CEA | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 5 precursor (Carcinoembryonic antigen) (CEA) (Meconium antigen 100) (CD66e antigen). | |||||
|
ESAM_MOUSE
|
||||||
| NC score | 0.062377 (rank : 42) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q925F2 | Gene names | Esam, Esam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
CXAR_MOUSE
|
||||||
| NC score | 0.061064 (rank : 43) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97792, O09052, Q3ULD0, Q91W66, Q99KG0, Q9DBJ8 | Gene names | Cxadr, Car | |||
|
Domain Architecture |

|
|||||
| Description | Coxsackievirus and adenovirus receptor homolog precursor (CAR) (mCAR). | |||||
|
LY9_HUMAN
|
||||||
| NC score | 0.059491 (rank : 44) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HBG7, Q14775, Q5VYI3, Q9H4N5, Q9NQ24 | Gene names | LY9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9) (CD229 antigen). | |||||
|
KV2B_MOUSE
|
||||||
| NC score | 0.058324 (rank : 45) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
ESAM_HUMAN
|
||||||
| NC score | 0.057554 (rank : 46) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
KV1W_HUMAN
|
||||||
| NC score | 0.056737 (rank : 47) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04431 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-I region Walker precursor. | |||||
|
CD7_MOUSE
|
||||||
| NC score | 0.056601 (rank : 48) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P50283 | Gene names | Cd7 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell antigen CD7 precursor. | |||||
|
SIRPD_HUMAN
|
||||||
| NC score | 0.055992 (rank : 49) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H106, Q5TFQ6 | Gene names | SIRPD, PTPNS1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein delta precursor (Protein tyrosine phosphatase non-receptor type substrate 1-like 2). | |||||
|
KV2F_HUMAN
|
||||||
| NC score | 0.053349 (rank : 50) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P06310 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-II region RPMI 6410 precursor. | |||||
|
CXAR_HUMAN
|
||||||
| NC score | 0.053263 (rank : 51) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78310, O00694, Q8WWT6, Q8WWT7, Q8WWT8, Q9UKV4 | Gene names | CXADR, CAR | |||
|
Domain Architecture |

|
|||||
| Description | Coxsackievirus and adenovirus receptor precursor (Coxsackievirus B- adenovirus receptor) (hCAR) (CVB3-binding protein) (HCVADR). | |||||
|
CD226_HUMAN
|
||||||
| NC score | 0.052382 (rank : 52) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15762 | Gene names | CD226, DNAM1 | |||
|
Domain Architecture |

|
|||||
| Description | CD226 antigen precursor (DNAX accessory molecule 1) (DNAM-1). | |||||
|
CD22_HUMAN
|
||||||
| NC score | 0.051663 (rank : 53) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
KV2A_MOUSE
|
||||||
| NC score | 0.051606 (rank : 54) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
JAM3_MOUSE
|
||||||
| NC score | 0.050944 (rank : 55) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8B7, Q8BT59, Q9D1M9, Q9EPK4 | Gene names | Jam3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctional adhesion molecule C precursor (JAM-C) (Junctional adhesion molecule 3) (JAM-3) (JAM-2). | |||||
|
KV1F_HUMAN
|
||||||
| NC score | 0.048526 (rank : 56) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P01598 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region EU. | |||||
|
KV1Y_HUMAN
|
||||||
| NC score | 0.046968 (rank : 57) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80362 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region WAT. | |||||
|
KV1E_HUMAN
|
||||||
| NC score | 0.046896 (rank : 58) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01597 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region DEE. | |||||
|
KV1Q_HUMAN
|
||||||
| NC score | 0.046825 (rank : 59) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01609 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Scw. | |||||
|
KV1D_HUMAN
|
||||||
| NC score | 0.046801 (rank : 60) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01596 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region CAR. | |||||
|
KV1L_HUMAN
|
||||||
| NC score | 0.045844 (rank : 61) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01604 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Kue. | |||||
|
KV5P_MOUSE
|
||||||
| NC score | 0.045420 (rank : 62) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01649 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-V regions (Anti-arsonate antibodies). | |||||
|
KV1G_HUMAN
|
||||||
| NC score | 0.044596 (rank : 63) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P01599 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Gal. | |||||
|
KV1P_HUMAN
|
||||||
| NC score | 0.044426 (rank : 64) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01608 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Roy. | |||||
|
IGSF2_HUMAN
|
||||||
| NC score | 0.043634 (rank : 65) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q93033, Q15856 | Gene names | IGSF2, CD101, EWI101, V7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin superfamily member 2 precursor (Glu-Trp-Ile EWI motif- containing protein 101) (EWI-101) (Cell surface glycoprotein V7) (CD101 antigen). | |||||
|
KV5R_MOUSE
|
||||||
| NC score | 0.041564 (rank : 66) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01651 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-V region EPC 109. | |||||
|
GI24_MOUSE
|
||||||
| NC score | 0.041112 (rank : 67) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D659, Q6KAT9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet receptor Gi24 precursor. | |||||
|
FGRL1_HUMAN
|
||||||
| NC score | 0.036474 (rank : 68) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
HPLN4_HUMAN
|
||||||
| NC score | 0.031832 (rank : 69) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.031614 (rank : 70) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
TYRO3_MOUSE
|
||||||
| NC score | 0.030218 (rank : 71) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55144, O09070, O09080, P70285, Q60752, Q62482, Q62483, Q62484, Q78E85, Q78E87 | Gene names | Tyro3, Dtk, Rse | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase DTK) (TK19-2) (Etk2/tyro3). | |||||
|
FCRLA_MOUSE
|
||||||
| NC score | 0.025759 (rank : 72) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
TYRO3_HUMAN
|
||||||
| NC score | 0.024260 (rank : 73) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06418, O14953, Q86VR3 | Gene names | TYRO3, BYK, DTK, RSE, SKY | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase SKY) (Tyrosine- protein kinase DTK) (Protein-tyrosine kinase byk). | |||||
|
HV46_MOUSE
|
||||||
| NC score | 0.016887 (rank : 74) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P01822 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig heavy chain V region MOPC 315 precursor. | |||||
|
HV12_MOUSE
|
||||||
| NC score | 0.009445 (rank : 75) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P01756 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig heavy chain V region MOPC 104E. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.006391 (rank : 76) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.005185 (rank : 77) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||