Please be patient as the page loads
|
CI078_HUMAN
|
||||||
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CI078_HUMAN
|
||||||
| θ value | 4.87785e-155 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CI078_MOUSE
|
||||||
| θ value | 7.54289e-148 (rank : 2) | NC score | 0.973983 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 3) | NC score | 0.141711 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.047722 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.101722 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.099772 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.110601 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.031115 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.128184 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.063757 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.075504 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.077871 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.079316 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.052782 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.045894 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.101809 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.059697 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.080582 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.068221 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.066338 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.093881 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.043432 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.052395 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.067664 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.055742 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.051751 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.050660 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.043602 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.037182 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.056585 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.046570 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.034883 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.062155 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.026095 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.055293 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.043670 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.065139 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.039609 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.069339 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RIT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.012767 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.057136 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.019047 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.013043 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.018967 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
GNL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.035700 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.049382 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.055626 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.041801 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.029351 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
NOL10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.034740 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.024101 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.029096 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.079067 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.049796 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.050396 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KAD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.022015 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |
|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.039239 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.029936 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.041923 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.009242 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CD017_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.052399 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q53FE4, Q6FI84, Q6IS77, Q9H0D9 | Gene names | C4orf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf17. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.051618 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.049789 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.054254 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.019070 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
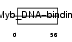
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.033718 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050694 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.055329 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.050820 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056429 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.062670 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.060392 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.051285 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.050911 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.051691 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SDS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053482 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H7L9, Q8N6H0, Q9H8D2 | Gene names | SUDS3, SDS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
SDS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.053198 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BR65, Q3UC92, Q6P6K1, Q7TNT0, Q8BRR7, Q8K5B4 | Gene names | Suds3, Sds3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.055686 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.055514 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.053103 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056018 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CI078_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.87785e-155 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CI078_MOUSE
|
||||||
| NC score | 0.973983 (rank : 2) | θ value | 7.54289e-148 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.141711 (rank : 3) | θ value | 0.00869519 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.128184 (rank : 4) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.110601 (rank : 5) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.101809 (rank : 6) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.101722 (rank : 7) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.099772 (rank : 8) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.093881 (rank : 9) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.080582 (rank : 10) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.079316 (rank : 11) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.079067 (rank : 12) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.077871 (rank : 13) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.075504 (rank : 14) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.069339 (rank : 15) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.068221 (rank : 16) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.067664 (rank : 17) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.066338 (rank : 18) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.065139 (rank : 19) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.063757 (rank : 20) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.062670 (rank : 21) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.062155 (rank : 22) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.060392 (rank : 23) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.059697 (rank : 24) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.057136 (rank : 25) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.056585 (rank : 26) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.056429 (rank : 27) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.056018 (rank : 28) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.055742 (rank : 29) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.055686 (rank : 30) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.055626 (rank : 31) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.055514 (rank : 32) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.055329 (rank : 33) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.055293 (rank : 34) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.054254 (rank : 35) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SDS3_HUMAN
|
||||||
| NC score | 0.053482 (rank : 36) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H7L9, Q8N6H0, Q9H8D2 | Gene names | SUDS3, SDS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
SDS3_MOUSE
|
||||||
| NC score | 0.053198 (rank : 37) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BR65, Q3UC92, Q6P6K1, Q7TNT0, Q8BRR7, Q8K5B4 | Gene names | Suds3, Sds3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.053103 (rank : 38) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.052782 (rank : 39) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CD017_HUMAN
|
||||||
| NC score | 0.052399 (rank : 40) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q53FE4, Q6FI84, Q6IS77, Q9H0D9 | Gene names | C4orf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf17. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.052395 (rank : 41) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.051751 (rank : 42) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.051691 (rank : 43) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.051618 (rank : 44) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.051285 (rank : 45) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.050911 (rank : 46) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.050820 (rank : 47) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.050694 (rank : 48) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.050660 (rank : 49) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.050396 (rank : 50) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.049796 (rank : 51) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.049789 (rank : 52) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.049382 (rank : 53) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.047722 (rank : 54) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.046570 (rank : 55) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.045894 (rank : 56) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.043670 (rank : 57) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.043602 (rank : 58) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.043432 (rank : 59) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.041923 (rank : 60) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.041801 (rank : 61) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.039609 (rank : 62) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.039239 (rank : 63) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.037182 (rank : 64) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GNL3_HUMAN
|
||||||
| NC score | 0.035700 (rank : 65) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BVP2, Q96SV6, Q96SV7, Q9UJY0 | Gene names | GNL3, E2IG3, NS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein-like 3 (Nucleolar GTP-binding protein 3) (Nucleostemin) (E2-induced gene 3-protein) (Novel nucleolar protein 47) (NNP47). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.034883 (rank : 66) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
NOL10_HUMAN
|
||||||
| NC score | 0.034740 (rank : 67) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.033718 (rank : 68) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.031115 (rank : 69) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.029936 (rank : 70) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.029351 (rank : 71) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.029096 (rank : 72) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.026095 (rank : 73) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.024101 (rank : 74) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
KAD2_HUMAN
|
||||||
| NC score | 0.022015 (rank : 75) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54819, Q16856, Q5TIF7, Q8TCY2, Q8TCY3 | Gene names | AK2, ADK2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 2, mitochondrial (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.019070 (rank : 76) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.019047 (rank : 77) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.018967 (rank : 78) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.013043 (rank : 79) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
RIT1_MOUSE
|
||||||
| NC score | 0.012767 (rank : 80) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.009242 (rank : 81) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||