Please be patient as the page loads
|
CCG6_MOUSE
|
||||||
| SwissProt Accessions | Q8VHW3 | Gene names | Cacng6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-6 subunit (Neuronal voltage- gated calcium channel gamma-6 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCG6_MOUSE
|
||||||
| θ value | 7.35691e-119 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHW3 | Gene names | Cacng6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-6 subunit (Neuronal voltage- gated calcium channel gamma-6 subunit). | |||||
|
CCG6_HUMAN
|
||||||
| θ value | 3.21347e-98 (rank : 2) | NC score | 0.996667 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXT2 | Gene names | CACNG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-6 subunit (Neuronal voltage- gated calcium channel gamma-6 subunit). | |||||
|
CCG1_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 3) | NC score | 0.847190 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06432 | Gene names | CACNG1, CACNLG | |||
|
Domain Architecture |
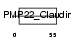
|
|||||
| Description | Voltage-dependent calcium channel gamma-1 subunit (Dihydropyridine- sensitive L-type, skeletal muscle calcium channel subunit gamma). | |||||
|
CCG1_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 4) | NC score | 0.800675 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70578 | Gene names | Cacng1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-1 subunit (Dihydropyridine- sensitive L-type, skeletal muscle calcium channel subunit gamma). | |||||
|
LMIP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.186500 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
LMIP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.180343 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56563, Q3TNV8, Q99PA6 | Gene names | Lim2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP17) (MP18) (MP19) (MP20). | |||||
|
CLD18_MOUSE
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.025302 (rank : 16) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56857, Q91ZY9, Q91ZZ0, Q91ZZ1 | Gene names | Cldn18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||
|
CCG4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.051758 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBN1 | Gene names | CACNG4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-4 subunit (Neuronal voltage- gated calcium channel gamma-4 subunit). | |||||
|
CCG4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.051768 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJV4 | Gene names | Cacng4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-4 subunit (Neuronal voltage- gated calcium channel gamma-4 subunit). | |||||
|
SSPN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.052406 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62147, Q60594 | Gene names | Sspn, Krag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcospan (K-ras oncogene-associated protein) (Kirsten-Ras-associated protein). | |||||
|
CCG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.056148 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UF02, Q8WXS7, Q9UHM3 | Gene names | CACNG5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
CCG5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.073104 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHW4 | Gene names | Cacng5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
SSPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.042858 (rank : 15) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14714 | Gene names | SSPN, KRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcospan (K-ras oncogene-associated protein) (Kirsten-ras-associated protein). | |||||
|
CCG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.057084 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60359 | Gene names | CACNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CCG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.063293 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJV5 | Gene names | Cacng3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
PERP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.050174 (rank : 14) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96FX8, Q8IWS3, Q8N1J6, Q8NC16, Q9H1C5, Q9H230 | Gene names | PERP, KCP1, KRTCAP1, PIGPC1, THW | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p53 apoptosis effector related to PMP-22 (Keratinocytes-associated protein 1) (KCP-1) (P53-induced protein PIGPC1) (Transmembrane protein THW). | |||||
|
CCG6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.35691e-119 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHW3 | Gene names | Cacng6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-6 subunit (Neuronal voltage- gated calcium channel gamma-6 subunit). | |||||
|
CCG6_HUMAN
|
||||||
| NC score | 0.996667 (rank : 2) | θ value | 3.21347e-98 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXT2 | Gene names | CACNG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-6 subunit (Neuronal voltage- gated calcium channel gamma-6 subunit). | |||||
|
CCG1_HUMAN
|
||||||
| NC score | 0.847190 (rank : 3) | θ value | 7.06379e-29 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06432 | Gene names | CACNG1, CACNLG | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-1 subunit (Dihydropyridine- sensitive L-type, skeletal muscle calcium channel subunit gamma). | |||||
|
CCG1_MOUSE
|
||||||
| NC score | 0.800675 (rank : 4) | θ value | 1.86321e-21 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O70578 | Gene names | Cacng1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent calcium channel gamma-1 subunit (Dihydropyridine- sensitive L-type, skeletal muscle calcium channel subunit gamma). | |||||
|
LMIP_HUMAN
|
||||||
| NC score | 0.186500 (rank : 5) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55344, Q9BXD0, Q9HAR5 | Gene names | LIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP18) (MP19) (MP20). | |||||
|
LMIP_MOUSE
|
||||||
| NC score | 0.180343 (rank : 6) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56563, Q3TNV8, Q99PA6 | Gene names | Lim2 | |||
|
Domain Architecture |

|
|||||
| Description | Lens fiber membrane intrinsic protein (MP17) (MP18) (MP19) (MP20). | |||||
|
CCG5_MOUSE
|
||||||
| NC score | 0.073104 (rank : 7) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHW4 | Gene names | Cacng5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
CCG3_MOUSE
|
||||||
| NC score | 0.063293 (rank : 8) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJV5 | Gene names | Cacng3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CCG3_HUMAN
|
||||||
| NC score | 0.057084 (rank : 9) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60359 | Gene names | CACNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-3 subunit (Neuronal voltage- gated calcium channel gamma-3 subunit). | |||||
|
CCG5_HUMAN
|
||||||
| NC score | 0.056148 (rank : 10) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UF02, Q8WXS7, Q9UHM3 | Gene names | CACNG5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-5 subunit (Neuronal voltage- gated calcium channel gamma-5 subunit). | |||||
|
SSPN_MOUSE
|
||||||
| NC score | 0.052406 (rank : 11) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62147, Q60594 | Gene names | Sspn, Krag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcospan (K-ras oncogene-associated protein) (Kirsten-Ras-associated protein). | |||||
|
CCG4_MOUSE
|
||||||
| NC score | 0.051768 (rank : 12) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJV4 | Gene names | Cacng4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-4 subunit (Neuronal voltage- gated calcium channel gamma-4 subunit). | |||||
|
CCG4_HUMAN
|
||||||
| NC score | 0.051758 (rank : 13) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBN1 | Gene names | CACNG4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent calcium channel gamma-4 subunit (Neuronal voltage- gated calcium channel gamma-4 subunit). | |||||
|
PERP_HUMAN
|
||||||
| NC score | 0.050174 (rank : 14) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96FX8, Q8IWS3, Q8N1J6, Q8NC16, Q9H1C5, Q9H230 | Gene names | PERP, KCP1, KRTCAP1, PIGPC1, THW | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p53 apoptosis effector related to PMP-22 (Keratinocytes-associated protein 1) (KCP-1) (P53-induced protein PIGPC1) (Transmembrane protein THW). | |||||
|
SSPN_HUMAN
|
||||||
| NC score | 0.042858 (rank : 15) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14714 | Gene names | SSPN, KRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcospan (K-ras oncogene-associated protein) (Kirsten-ras-associated protein). | |||||
|
CLD18_MOUSE
|
||||||
| NC score | 0.025302 (rank : 16) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56857, Q91ZY9, Q91ZZ0, Q91ZZ1 | Gene names | Cldn18 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-18. | |||||