Please be patient as the page loads
|
CANT1_MOUSE
|
||||||
| SwissProt Accessions | Q8VCF1, Q8C3R8, Q8C4T6, Q9D3F2, Q9D9R1 | Gene names | Cant1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CANT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996977 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVQ1, Q7Z2J7, Q8NG05, Q8NHP0, Q9BSD5 | Gene names | CANT1, SHAPY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog) (Putative NF-kappa-B-activating protein 107) (Putative MAPK-activating protein PM09). | |||||
|
CANT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCF1, Q8C3R8, Q8C4T6, Q9D3F2, Q9D9R1 | Gene names | Cant1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog). | |||||
|
TRI36_MOUSE
|
||||||
| θ value | 0.163984 (rank : 3) | NC score | 0.053907 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
TRI36_HUMAN
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.046332 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
AP2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 5) | NC score | 0.034056 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92481, Q5JYX6, Q9NQ63, Q9NU99, Q9UJI7, Q9Y214, Q9Y3K3 | Gene names | TFAP2B | |||
|
Domain Architecture |
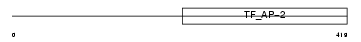
|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
AP2B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.034021 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61313 | Gene names | Tfap2b, Tcfap2b | |||
|
Domain Architecture |
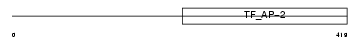
|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
DPYL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.028285 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16555, O00424 | Gene names | DPYSL2, CRMP2 | |||
|
Domain Architecture |
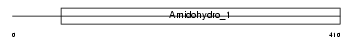
|
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (Collapsin response mediator protein 2) (CRMP-2) (N2A3). | |||||
|
DPYL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 8) | NC score | 0.028271 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08553, Q6P5D0 | Gene names | Dpysl2, Crmp2, Ulip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (ULIP 2 protein). | |||||
|
PROS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.015029 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08761, P43483 | Gene names | Pros1, Pros | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
DPYS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.026761 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14117 | Gene names | DPYS | |||
|
Domain Architecture |
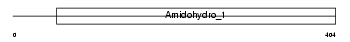
|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
EMIL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.011894 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXX0, Q8NBH3, Q96JQ4 | Gene names | EMILIN2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Protein FOAP-10). | |||||
|
PCDH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.006901 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
CANT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCF1, Q8C3R8, Q8C4T6, Q9D3F2, Q9D9R1 | Gene names | Cant1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog). | |||||
|
CANT1_HUMAN
|
||||||
| NC score | 0.996977 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVQ1, Q7Z2J7, Q8NG05, Q8NHP0, Q9BSD5 | Gene names | CANT1, SHAPY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Soluble calcium-activated nucleotidase 1 (EC 3.6.1.6) (SCAN-1) (Apyrase homolog) (Putative NF-kappa-B-activating protein 107) (Putative MAPK-activating protein PM09). | |||||
|
TRI36_MOUSE
|
||||||
| NC score | 0.053907 (rank : 3) | θ value | 0.163984 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WG7, Q7TNM1 | Gene names | Trim36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Haprin) (Acrosome RBCC protein). | |||||
|
TRI36_HUMAN
|
||||||
| NC score | 0.046332 (rank : 4) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQ86 | Gene names | TRIM36, RBCC728, RNF98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 36 (Zinc-binding protein Rbcc728) (RING finger protein 98). | |||||
|
AP2B_HUMAN
|
||||||
| NC score | 0.034056 (rank : 5) | θ value | 2.36792 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92481, Q5JYX6, Q9NQ63, Q9NU99, Q9UJI7, Q9Y214, Q9Y3K3 | Gene names | TFAP2B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
AP2B_MOUSE
|
||||||
| NC score | 0.034021 (rank : 6) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61313 | Gene names | Tfap2b, Tcfap2b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
DPYL2_HUMAN
|
||||||
| NC score | 0.028285 (rank : 7) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16555, O00424 | Gene names | DPYSL2, CRMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (Collapsin response mediator protein 2) (CRMP-2) (N2A3). | |||||
|
DPYL2_MOUSE
|
||||||
| NC score | 0.028271 (rank : 8) | θ value | 2.36792 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08553, Q6P5D0 | Gene names | Dpysl2, Crmp2, Ulip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropyrimidinase-related protein 2 (DRP-2) (ULIP 2 protein). | |||||
|
DPYS_HUMAN
|
||||||
| NC score | 0.026761 (rank : 9) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14117 | Gene names | DPYS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydropyrimidinase (EC 3.5.2.2) (DHPase) (Hydantoinase) (DHP). | |||||
|
PROS_MOUSE
|
||||||
| NC score | 0.015029 (rank : 10) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08761, P43483 | Gene names | Pros1, Pros | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein S precursor. | |||||
|
EMIL2_HUMAN
|
||||||
| NC score | 0.011894 (rank : 11) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXX0, Q8NBH3, Q96JQ4 | Gene names | EMILIN2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Protein FOAP-10). | |||||
|
PCDH7_HUMAN
|
||||||
| NC score | 0.006901 (rank : 12) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||