Please be patient as the page loads
|
ANX13_MOUSE
|
||||||
| SwissProt Accessions | Q99JG3 | Gene names | Anxa13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Annexin A13 (Annexin XIII). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ANX13_MOUSE
|
||||||
| θ value | 7.7336e-177 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99JG3 | Gene names | Anxa13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Annexin A13 (Annexin XIII). | |||||
|
ANX13_HUMAN
|
||||||
| θ value | 3.27187e-151 (rank : 2) | NC score | 0.998093 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27216, Q9BQR5 | Gene names | ANXA13, ANX13 | |||
|
Domain Architecture |
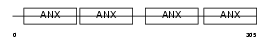
|
|||||
| Description | Annexin A13 (Annexin XIII) (Annexin, intestine-specific) (ISA). | |||||
|
ANXA7_MOUSE
|
||||||
| θ value | 2.64101e-84 (rank : 3) | NC score | 0.965869 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |
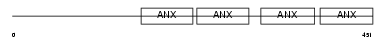
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
ANXA7_HUMAN
|
||||||
| θ value | 1.31072e-83 (rank : 4) | NC score | 0.966841 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |
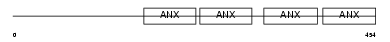
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 8.22888e-78 (rank : 5) | NC score | 0.970728 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
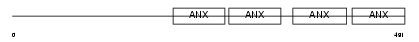
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
ANXA8_HUMAN
|
||||||
| θ value | 8.22888e-78 (rank : 6) | NC score | 0.988134 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13928, Q9BT34 | Gene names | ANXA8, ANX8 | |||
|
Domain Architecture |
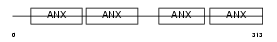
|
|||||
| Description | Annexin A8 (Annexin VIII) (Vascular anticoagulant-beta) (VAC-beta). | |||||
|
ANXA4_HUMAN
|
||||||
| θ value | 1.07473e-77 (rank : 7) | NC score | 0.989316 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09525, Q96F33, Q9BWK1 | Gene names | ANXA4, ANX4 | |||
|
Domain Architecture |
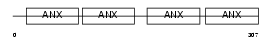
|
|||||
| Description | Annexin A4 (Annexin IV) (Lipocortin IV) (Endonexin I) (Chromobindin-4) (Protein II) (P32.5) (Placental anticoagulant protein II) (PAP-II) (PP4-X) (35-beta calcimedin) (Carbohydrate-binding protein P33/P41) (P33/41). | |||||
|
ANXA4_MOUSE
|
||||||
| θ value | 1.18825e-76 (rank : 8) | NC score | 0.989322 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97429 | Gene names | Anxa4, Anx4 | |||
|
Domain Architecture |
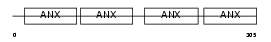
|
|||||
| Description | Annexin A4 (Annexin IV). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 2.64714e-76 (rank : 9) | NC score | 0.968339 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
ANXA6_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 10) | NC score | 0.979058 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08133, Q6ZT79 | Gene names | ANXA6, ANX6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
ANXA6_MOUSE
|
||||||
| θ value | 5.51964e-74 (rank : 11) | NC score | 0.978699 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14824 | Gene names | Anxa6, Anx6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
ANXA8_MOUSE
|
||||||
| θ value | 1.04096e-72 (rank : 12) | NC score | 0.987276 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35640 | Gene names | Anxa8, Anx8 | |||
|
Domain Architecture |
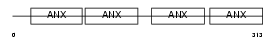
|
|||||
| Description | Annexin A8 (Annexin VIII). | |||||
|
ANXA5_MOUSE
|
||||||
| θ value | 1.15091e-71 (rank : 13) | NC score | 0.987715 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48036, Q99LA1 | Gene names | Anxa5, Anx5 | |||
|
Domain Architecture |
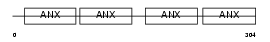
|
|||||
| Description | Annexin A5 (Annexin V) (Lipocortin V) (Endonexin II) (Calphobindin I) (CBP-I) (Placental anticoagulant protein I) (PAP-I) (PP4) (Thromboplastin inhibitor) (Vascular anticoagulant-alpha) (VAC-alpha) (Anchorin CII). | |||||
|
ANXA5_HUMAN
|
||||||
| θ value | 4.37348e-71 (rank : 14) | NC score | 0.987709 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08758, Q6FI16, Q8WV69 | Gene names | ANXA5, ANX5, ENX2, PP4 | |||
|
Domain Architecture |
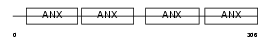
|
|||||
| Description | Annexin A5 (Annexin V) (Lipocortin V) (Endonexin II) (Calphobindin I) (CBP-I) (Placental anticoagulant protein I) (PAP-I) (PP4) (Thromboplastin inhibitor) (Vascular anticoagulant-alpha) (VAC-alpha) (Anchorin CII). | |||||
|
ANXA3_MOUSE
|
||||||
| θ value | 1.40689e-69 (rank : 15) | NC score | 0.986658 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35639 | Gene names | Anxa3, Anx3 | |||
|
Domain Architecture |
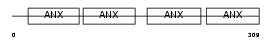
|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin). | |||||
|
ANXA3_HUMAN
|
||||||
| θ value | 3.4653e-68 (rank : 16) | NC score | 0.986651 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12429, Q6LET2 | Gene names | ANXA3, ANX3 | |||
|
Domain Architecture |
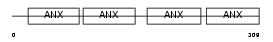
|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin) (Inositol 1,2-cyclic phosphate 2-phosphohydrolase). | |||||
|
ANXA1_HUMAN
|
||||||
| θ value | 1.71981e-67 (rank : 17) | NC score | 0.985000 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04083 | Gene names | ANXA1, ANX1, LPC1 | |||
|
Domain Architecture |
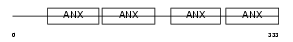
|
|||||
| Description | Annexin A1 (Annexin I) (Lipocortin I) (Calpactin II) (Chromobindin-9) (p35) (Phospholipase A2 inhibitory protein). | |||||
|
ANXA1_MOUSE
|
||||||
| θ value | 1.71981e-67 (rank : 18) | NC score | 0.984546 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10107 | Gene names | Anxa1, Anx1, Lpc-1, Lpc1 | |||
|
Domain Architecture |
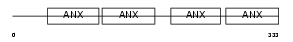
|
|||||
| Description | Annexin A1 (Annexin I) (Lipocortin I) (Calpactin II) (Chromobindin-9) (p35) (Phospholipase A2 inhibitory protein). | |||||
|
ANXA2_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 19) | NC score | 0.983263 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07355, Q96DD5 | Gene names | ANXA2, ANX2 | |||
|
Domain Architecture |
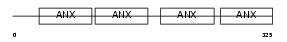
|
|||||
| Description | Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulant protein IV) (PAP-IV). | |||||
|
ANXA2_MOUSE
|
||||||
| θ value | 5.53243e-66 (rank : 20) | NC score | 0.983437 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07356 | Gene names | Anxa2, Anx2, Cal1h | |||
|
Domain Architecture |
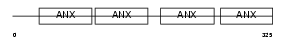
|
|||||
| Description | Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulant protein IV) (PAP-IV). | |||||
|
ANX10_MOUSE
|
||||||
| θ value | 1.66964e-54 (rank : 21) | NC score | 0.978630 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QZ10 | Gene names | Anxa10 | |||
|
Domain Architecture |
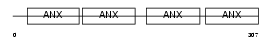
|
|||||
| Description | Annexin A10. | |||||
|
ANX10_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 22) | NC score | 0.977730 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ72, Q96IQ5, Q9UJV4 | Gene names | ANXA10, ANX14 | |||
|
Domain Architecture |
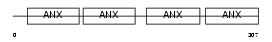
|
|||||
| Description | Annexin A10 (Annexin-14). | |||||
|
ANXA9_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 23) | NC score | 0.966239 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O76027, Q5SZF1, Q6FI55, Q9BS00, Q9HBJ6 | Gene names | ANXA9, ANX31 | |||
|
Domain Architecture |
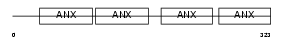
|
|||||
| Description | Annexin A9 (Annexin-31) (Annexin XXXI) (Pemphaxin). | |||||
|
ANXA9_MOUSE
|
||||||
| θ value | 6.81017e-40 (rank : 24) | NC score | 0.966281 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHQ0, Q9CQS1 | Gene names | Anxa9 | |||
|
Domain Architecture |
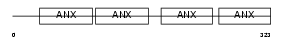
|
|||||
| Description | Annexin A9 (Annexin-31) (Annexin XXXI). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.013954 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.026094 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.031446 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.004767 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
TRI12_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.004927 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
FES_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | -0.002604 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.015348 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
RAD21_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.022339 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60216, Q15001, Q99568 | Gene names | RAD21, HR21, KIAA0078, NXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (hHR21) (Nuclear matrix protein 1) (NXP-1) (SCC1 homolog). | |||||
|
1433Z_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.008102 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63104, P29213, P29312, Q32P43, Q5XJ08, Q6GPI2, Q6IN74, Q6NUR9, Q6P3U9, Q86V33 | Gene names | YWHAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.006314 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
RAD21_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.020254 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61550, P70219, Q91VB9, Q9DBU4 | Gene names | Rad21, Hr21 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (Pokeweed agglutinin- binding protein 29) (PW29) (SCC1 homolog). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.002822 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.003129 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.013466 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.006887 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.009390 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
1433Z_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.006343 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63101, P35215, P70197, P97286, Q3TSF1, Q5EBQ1 | Gene names | Ywhaz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1) (SEZ-2). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.004860 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.006384 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.000852 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.003602 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.002077 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.001754 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.002323 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.001757 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TBC12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.008757 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
ANX13_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.7336e-177 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99JG3 | Gene names | Anxa13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Annexin A13 (Annexin XIII). | |||||
|
ANX13_HUMAN
|
||||||
| NC score | 0.998093 (rank : 2) | θ value | 3.27187e-151 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27216, Q9BQR5 | Gene names | ANXA13, ANX13 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A13 (Annexin XIII) (Annexin, intestine-specific) (ISA). | |||||
|
ANXA4_MOUSE
|
||||||
| NC score | 0.989322 (rank : 3) | θ value | 1.18825e-76 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97429 | Gene names | Anxa4, Anx4 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A4 (Annexin IV). | |||||
|
ANXA4_HUMAN
|
||||||
| NC score | 0.989316 (rank : 4) | θ value | 1.07473e-77 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09525, Q96F33, Q9BWK1 | Gene names | ANXA4, ANX4 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A4 (Annexin IV) (Lipocortin IV) (Endonexin I) (Chromobindin-4) (Protein II) (P32.5) (Placental anticoagulant protein II) (PAP-II) (PP4-X) (35-beta calcimedin) (Carbohydrate-binding protein P33/P41) (P33/41). | |||||
|
ANXA8_HUMAN
|
||||||
| NC score | 0.988134 (rank : 5) | θ value | 8.22888e-78 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13928, Q9BT34 | Gene names | ANXA8, ANX8 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A8 (Annexin VIII) (Vascular anticoagulant-beta) (VAC-beta). | |||||
|
ANXA5_MOUSE
|
||||||
| NC score | 0.987715 (rank : 6) | θ value | 1.15091e-71 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48036, Q99LA1 | Gene names | Anxa5, Anx5 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A5 (Annexin V) (Lipocortin V) (Endonexin II) (Calphobindin I) (CBP-I) (Placental anticoagulant protein I) (PAP-I) (PP4) (Thromboplastin inhibitor) (Vascular anticoagulant-alpha) (VAC-alpha) (Anchorin CII). | |||||
|
ANXA5_HUMAN
|
||||||
| NC score | 0.987709 (rank : 7) | θ value | 4.37348e-71 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08758, Q6FI16, Q8WV69 | Gene names | ANXA5, ANX5, ENX2, PP4 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A5 (Annexin V) (Lipocortin V) (Endonexin II) (Calphobindin I) (CBP-I) (Placental anticoagulant protein I) (PAP-I) (PP4) (Thromboplastin inhibitor) (Vascular anticoagulant-alpha) (VAC-alpha) (Anchorin CII). | |||||
|
ANXA8_MOUSE
|
||||||
| NC score | 0.987276 (rank : 8) | θ value | 1.04096e-72 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35640 | Gene names | Anxa8, Anx8 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A8 (Annexin VIII). | |||||
|
ANXA3_MOUSE
|
||||||
| NC score | 0.986658 (rank : 9) | θ value | 1.40689e-69 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35639 | Gene names | Anxa3, Anx3 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin). | |||||
|
ANXA3_HUMAN
|
||||||
| NC score | 0.986651 (rank : 10) | θ value | 3.4653e-68 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12429, Q6LET2 | Gene names | ANXA3, ANX3 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A3 (Annexin III) (Lipocortin III) (Placental anticoagulant protein III) (PAP-III) (35-alpha calcimedin) (Inositol 1,2-cyclic phosphate 2-phosphohydrolase). | |||||
|
ANXA1_HUMAN
|
||||||
| NC score | 0.985000 (rank : 11) | θ value | 1.71981e-67 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04083 | Gene names | ANXA1, ANX1, LPC1 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A1 (Annexin I) (Lipocortin I) (Calpactin II) (Chromobindin-9) (p35) (Phospholipase A2 inhibitory protein). | |||||
|
ANXA1_MOUSE
|
||||||
| NC score | 0.984546 (rank : 12) | θ value | 1.71981e-67 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10107 | Gene names | Anxa1, Anx1, Lpc-1, Lpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A1 (Annexin I) (Lipocortin I) (Calpactin II) (Chromobindin-9) (p35) (Phospholipase A2 inhibitory protein). | |||||
|
ANXA2_MOUSE
|
||||||
| NC score | 0.983437 (rank : 13) | θ value | 5.53243e-66 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07356 | Gene names | Anxa2, Anx2, Cal1h | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulant protein IV) (PAP-IV). | |||||
|
ANXA2_HUMAN
|
||||||
| NC score | 0.983263 (rank : 14) | θ value | 1.90147e-66 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07355, Q96DD5 | Gene names | ANXA2, ANX2 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A2 (Annexin II) (Lipocortin II) (Calpactin I heavy chain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulant protein IV) (PAP-IV). | |||||
|
ANXA6_HUMAN
|
||||||
| NC score | 0.979058 (rank : 15) | θ value | 1.45253e-74 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08133, Q6ZT79 | Gene names | ANXA6, ANX6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
ANXA6_MOUSE
|
||||||
| NC score | 0.978699 (rank : 16) | θ value | 5.51964e-74 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14824 | Gene names | Anxa6, Anx6 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A6 (Annexin VI) (Lipocortin VI) (P68) (P70) (Protein III) (Chromobindin-20) (67 kDa calelectrin) (Calphobindin-II) (CPB-II). | |||||
|
ANX10_MOUSE
|
||||||
| NC score | 0.978630 (rank : 17) | θ value | 1.66964e-54 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QZ10 | Gene names | Anxa10 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A10. | |||||
|
ANX10_HUMAN
|
||||||
| NC score | 0.977730 (rank : 18) | θ value | 1.56274e-52 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ72, Q96IQ5, Q9UJV4 | Gene names | ANXA10, ANX14 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A10 (Annexin-14). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.970728 (rank : 19) | θ value | 8.22888e-78 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.968339 (rank : 20) | θ value | 2.64714e-76 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
ANXA7_HUMAN
|
||||||
| NC score | 0.966841 (rank : 21) | θ value | 1.31072e-83 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
ANXA9_MOUSE
|
||||||
| NC score | 0.966281 (rank : 22) | θ value | 6.81017e-40 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHQ0, Q9CQS1 | Gene names | Anxa9 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A9 (Annexin-31) (Annexin XXXI). | |||||
|
ANXA9_HUMAN
|
||||||
| NC score | 0.966239 (rank : 23) | θ value | 5.21438e-40 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O76027, Q5SZF1, Q6FI55, Q9BS00, Q9HBJ6 | Gene names | ANXA9, ANX31 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A9 (Annexin-31) (Annexin XXXI) (Pemphaxin). | |||||
|
ANXA7_MOUSE
|
||||||
| NC score | 0.965869 (rank : 24) | θ value | 2.64101e-84 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q07076 | Gene names | Anxa7, Anx7 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.031446 (rank : 25) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.026094 (rank : 26) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
RAD21_HUMAN
|
||||||
| NC score | 0.022339 (rank : 27) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60216, Q15001, Q99568 | Gene names | RAD21, HR21, KIAA0078, NXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (hHR21) (Nuclear matrix protein 1) (NXP-1) (SCC1 homolog). | |||||
|
RAD21_MOUSE
|
||||||
| NC score | 0.020254 (rank : 28) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61550, P70219, Q91VB9, Q9DBU4 | Gene names | Rad21, Hr21 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand-break repair protein rad21 homolog (Pokeweed agglutinin- binding protein 29) (PW29) (SCC1 homolog). | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.015348 (rank : 29) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.013954 (rank : 30) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.013466 (rank : 31) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.009390 (rank : 32) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
TBC12_HUMAN
|
||||||
| NC score | 0.008757 (rank : 33) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
1433Z_HUMAN
|
||||||
| NC score | 0.008102 (rank : 34) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63104, P29213, P29312, Q32P43, Q5XJ08, Q6GPI2, Q6IN74, Q6NUR9, Q6P3U9, Q86V33 | Gene names | YWHAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.006887 (rank : 35) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.006384 (rank : 36) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
1433Z_MOUSE
|
||||||
| NC score | 0.006343 (rank : 37) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63101, P35215, P70197, P97286, Q3TSF1, Q5EBQ1 | Gene names | Ywhaz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 14-3-3 protein zeta/delta (Protein kinase C inhibitor protein 1) (KCIP-1) (SEZ-2). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.006314 (rank : 38) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TRI12_MOUSE
|
||||||
| NC score | 0.004927 (rank : 39) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.004860 (rank : 40) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.004767 (rank : 41) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.003602 (rank : 42) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.003129 (rank : 43) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.002822 (rank : 44) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.002323 (rank : 45) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.002077 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.001757 (rank : 47) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.001754 (rank : 48) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.000852 (rank : 49) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FES_HUMAN
|
||||||
| NC score | -0.002604 (rank : 50) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07332 | Gene names | FES, FPS | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||