Please be patient as the page loads
|
AGM1_HUMAN
|
||||||
| SwissProt Accessions | O95394, Q5JWR4, Q96J46, Q9H8G5, Q9NS94, Q9NTT6, Q9UFV5, Q9UIY2 | Gene names | PGM3, AGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AGM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95394, Q5JWR4, Q96J46, Q9H8G5, Q9NS94, Q9NTT6, Q9UFV5, Q9UIY2 | Gene names | PGM3, AGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3). | |||||
|
AGM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997591 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CYR6, Q543F9 | Gene names | Pgm3, Agm1, Pgm-3 | |||
|
Domain Architecture |
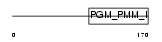
|
|||||
| Description | Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3). | |||||
|
PGM2L_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 3) | NC score | 0.157296 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CAA7 | Gene names | Pgm2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2-like 1 (EC 5.4.2.2). | |||||
|
PGM2L_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.155133 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PCE3, Q96MQ7, Q9UIK3 | Gene names | PGM2L1, BM32A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2-like 1 (EC 5.4.2.2) (PMMLP). | |||||
|
PGM2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.146652 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96G03, Q53FP5, Q5QTR0, Q9H0P9, Q9NV22 | Gene names | PGM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2 (EC 5.4.2.2) (Glucose phosphomutase 2) (PGM 2). | |||||
|
PGM2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.146538 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TSV4, Q8K0P7, Q9CRS8 | Gene names | Pgm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2 (EC 5.4.2.2) (Glucose phosphomutase 2) (PGM 2). | |||||
|
CO3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.020162 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
REST_MOUSE
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.005872 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
GSCR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.038567 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZM5, Q9BTC6, Q9HAX6, Q9NPP1, Q9NPR4, Q9UFI2 | Gene names | GLTSCR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 2 protein (p60). | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.013243 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
AGM1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95394, Q5JWR4, Q96J46, Q9H8G5, Q9NS94, Q9NTT6, Q9UFV5, Q9UIY2 | Gene names | PGM3, AGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3). | |||||
|
AGM1_MOUSE
|
||||||
| NC score | 0.997591 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CYR6, Q543F9 | Gene names | Pgm3, Agm1, Pgm-3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoacetylglucosamine mutase (EC 5.4.2.3) (PAGM) (Acetylglucosamine phosphomutase) (N-acetylglucosamine-phosphate mutase) (Phosphoglucomutase 3). | |||||
|
PGM2L_MOUSE
|
||||||
| NC score | 0.157296 (rank : 3) | θ value | 0.0431538 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CAA7 | Gene names | Pgm2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2-like 1 (EC 5.4.2.2). | |||||
|
PGM2L_HUMAN
|
||||||
| NC score | 0.155133 (rank : 4) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PCE3, Q96MQ7, Q9UIK3 | Gene names | PGM2L1, BM32A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2-like 1 (EC 5.4.2.2) (PMMLP). | |||||
|
PGM2_HUMAN
|
||||||
| NC score | 0.146652 (rank : 5) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96G03, Q53FP5, Q5QTR0, Q9H0P9, Q9NV22 | Gene names | PGM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2 (EC 5.4.2.2) (Glucose phosphomutase 2) (PGM 2). | |||||
|
PGM2_MOUSE
|
||||||
| NC score | 0.146538 (rank : 6) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TSV4, Q8K0P7, Q9CRS8 | Gene names | Pgm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoglucomutase-2 (EC 5.4.2.2) (Glucose phosphomutase 2) (PGM 2). | |||||
|
GSCR2_HUMAN
|
||||||
| NC score | 0.038567 (rank : 7) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZM5, Q9BTC6, Q9HAX6, Q9NPP1, Q9NPR4, Q9UFI2 | Gene names | GLTSCR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 2 protein (p60). | |||||
|
CO3_HUMAN
|
||||||
| NC score | 0.020162 (rank : 8) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.013243 (rank : 9) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.005872 (rank : 10) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||